Difference between revisions of "Preparing an AMBER topology file for a protein plus ligand system"
import>Csw34 |
import>Csw34 |
||
| Line 15: | Line 15: | ||
Starting point: [[Media:2hu4.txt | 2hu4.pdb]] |
Starting point: [[Media:2hu4.txt | 2hu4.pdb]] |
||
| − | ===== |
+ | ===== Removing Hs from the protein ===== |
As [[AMBER]] uses an all-atop representation, we need to allow it to add in Hs as it sees fit when you generate the topology file. This is a problem if you already have Hs in the structure so for any part which you are going to load parameters for from the AMBER libraries, you must remove ALL H. This is best done using a script or a series of piped commands. For example, first, you find which types of H you have in the molecule using: |
As [[AMBER]] uses an all-atop representation, we need to allow it to add in Hs as it sees fit when you generate the topology file. This is a problem if you already have Hs in the structure so for any part which you are going to load parameters for from the AMBER libraries, you must remove ALL H. This is best done using a script or a series of piped commands. For example, first, you find which types of H you have in the molecule using: |
||
| Line 49: | Line 49: | ||
Don't worry about the atom numbering being messed up, we'll deal with that later! |
Don't worry about the atom numbering being messed up, we'll deal with that later! |
||
| − | ===== |
+ | ===== Adding H to the ligand ===== |
In my pdb file, there is a residue ''OTV''' corresponding to the drug molecule. When I generated parameters in AMBER, it had a complete set of Hs, and it's obvious from the PDB that it doesn't any more. This is a problem! We will need to add them back in using Molfacture, a tool in VMD. You will need the latest version of VMD to do this so either download and compile it yourself (which is pretty easy - honest!), or use my binary located at |
In my pdb file, there is a residue ''OTV''' corresponding to the drug molecule. When I generated parameters in AMBER, it had a complete set of Hs, and it's obvious from the PDB that it doesn't any more. This is a problem! We will need to add them back in using Molfacture, a tool in VMD. You will need the latest version of VMD to do this so either download and compile it yourself (which is pretty easy - honest!), or use my binary located at |
||
| Line 61: | Line 61: | ||
'''NOTE''':The coordinates have been preserved from the original crystal structure position of the ligand. This will turn out to be very useful later on! |
'''NOTE''':The coordinates have been preserved from the original crystal structure position of the ligand. This will turn out to be very useful later on! |
||
| − | ===== |
+ | ===== Checking for consistant atom naming ===== |
This is a vital step as when you parametrised your ligand originally, each atom was assigned a name from the generalised forcefield. Unless the atoms names in your pdb match with those in the AMBER ''prepin'' file, you're not going to be able to make a topology or coordinate file to use with AMBER. Unfortunately, this just requires some manual editing, there's no way around it! |
This is a vital step as when you parametrised your ligand originally, each atom was assigned a name from the generalised forcefield. Unless the atoms names in your pdb match with those in the AMBER ''prepin'' file, you're not going to be able to make a topology or coordinate file to use with AMBER. Unfortunately, this just requires some manual editing, there's no way around it! |
||
Revision as of 17:24, 12 May 2008
When you want to use AMBER, either on its own to run MD, or interfaced with any of the group software (GMIN or OPTIM), you need to have two files at your disposal. These are the topology file (which describes the connectivity of atoms, angles etc) and the coordinate file (which shockingly defines the coordinates of the atoms in your system).
For big systems, generating these files can take a bit of time, and the difficulty increases when you are trying to simulate something for which parameters do not exist in the AMBER libraries i.e. non-standard amino acids, or, more usually, a system with a novel ligand (for example a drug molecule). This tutorial will deal with a big system, hopefully covering all bases. You will probably find the process much less arduous when you are dealing with something smaller and I will try to point out where I am deviating from what you would need to do if you only had say a protein composed of standard amino acids.
I will also assume that you have already generated parameters (AMBER prepin and frcmod files) for any novel ligand/molecule that is present in your system. Info on how to do this can be found linked from the AMBER page.
Right! Lets begin:
Step 1: Preparing your pdb file
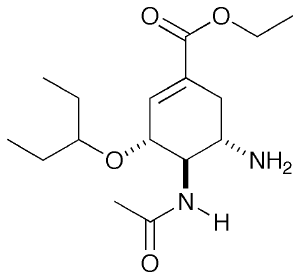
For this tutorial, I will be looking at neuraminidase (NA), an Influenza virus surface protein whose natural substrate is sialic acid. There are a few drugs which target this molecule as competitive inhibitors and one well known example is Tamiflu. Shown on the right is in fact the pro-drug form which you take. In the body, the ester is hydrolysed to a carboxylic acid and this is the form I will be looking at.
The starting point is a crystal structure of Tamiflu (oseltamivir) bound to NA which I obtained from the Protein Data Bank. NA is actually a tetramer so to cut down the simulation time, I will only consider the monomer and therefore I removed all but one copy from the PDB file, just by deleting the entries for the atoms.
Starting point: 2hu4.pdb
Removing Hs from the protein
As AMBER uses an all-atop representation, we need to allow it to add in Hs as it sees fit when you generate the topology file. This is a problem if you already have Hs in the structure so for any part which you are going to load parameters for from the AMBER libraries, you must remove ALL H. This is best done using a script or a series of piped commands. For example, first, you find which types of H you have in the molecule using:
grep H 2hu4.pdb | more
You'll get something like this:
ATOM 2 H1 VAL 1 43.584 25.333 13.539 1.00 0.00 ATOM 3 H2 VAL 1 44.124 26.003 14.929 1.00 0.00 ATOM 4 H3 VAL 1 42.554 25.613 14.779 1.00 0.00 ATOM 12 H LYS 2 43.614 29.813 13.829 1.00 0.00 ATOM 19 HZ1 LYS 2 48.594 30.403 12.569 1.00 0.00 ATOM 20 HZ2 LYS 2 47.324 29.383 12.669 1.00 0.00 ATOM 21 HZ3 LYS 2 47.194 30.783 11.829 1.00 0.00 ATOM 25 H LEU 3 41.774 31.293 17.769 1.00 0.00 ATOM 34 H ALA 4 38.304 32.193 15.399 1.00 0.00 ATOM 40 H GLY 5 37.974 36.783 16.069 1.00 0.00 ATOM 45 H ASN 6 35.304 36.683 15.349 1.00 0.00 ATOM 51 HD21 ASN 6 34.094 36.233 14.939 1.00 0.00 ATOM 52 HD22 ASN 6 33.744 35.023 13.739 1.00 0.00 ...
The task now is to find a pattern which will let you remove each type of H in turn using grep -v. i.e. I can remove the first three types of H using:
grep -v ' H[1-3]' 2hu4.pdb
NOTE: I've got a space before the H there. This is so that I don't accidentally remove lines with NH1 on for example. A good method is just to do a normal grep for the pattern that you plan on using to check that you won't catch any atoms you don't intend to! Once you've found a patten that covers all types of H in your molecule, you need to string them together and redirect the output to a new pdb file e.g.
grep -v ' H[1-3] ' 2hu4.pdb | grep -v H[Z,D,H,G] | grep -v ' HE' | grep -v ' H ' > 2hu4_noH.pdb
The results is here: 2hu4_noH.pdb
Don't worry about the atom numbering being messed up, we'll deal with that later!
Adding H to the ligand
In my pdb file, there is a residue OTV' corresponding to the drug molecule. When I generated parameters in AMBER, it had a complete set of Hs, and it's obvious from the PDB that it doesn't any more. This is a problem! We will need to add them back in using Molfacture, a tool in VMD. You will need the latest version of VMD to do this so either download and compile it yourself (which is pretty easy - honest!), or use my binary located at
clust:~csw34/bin/vmd
Using Molfacture to do this is explained in a seperate tutorial (as it's a bit long winded) in the VMD section. Once you have added all the Hs required and ensured that the charge on the molecule is what you require (-1 in this case due to a COO- group), you need to save the ligand you are editing as a new PDB using File>Save and changing the type to .pdb. The result is linked below:
Ligand plus Hs from VMD: vmdligand.pdb
NOTE:The coordinates have been preserved from the original crystal structure position of the ligand. This will turn out to be very useful later on!
Checking for consistant atom naming
This is a vital step as when you parametrised your ligand originally, each atom was assigned a name from the generalised forcefield. Unless the atoms names in your pdb match with those in the AMBER prepin file, you're not going to be able to make a topology or coordinate file to use with AMBER. Unfortunately, this just requires some manual editing, there's no way around it!