Preparing an AMBER topology file for a protein plus ligand system
When you want to use AMBER, either on its own to run MD, or interfaced with any of the group software (GMIN, OPTIM or PATHSAMPLE), you need to have two files at your disposal. These are the topology file (which describes the atom types, connectivity of atoms, force field parameters for angles, etc.) and the coordinate file (which shockingly defines the coordinates of the atoms in your system).
For big systems, generating these files can take a bit of time, and the difficulty increases when you are trying to simulate something for which parameters do not exist in the AMBER libraries i.e. non-standard amino acids, or, more usually, a system with a novel ligand (for example a drug molecule). This tutorial will deal with a big system, hopefully covering all bases. You will probably find the process much less arduous when you are dealing with something smaller and I will try to point out where I am deviating from what you would need to do if you only had say a protein composed of standard amino acids.
I will also assume that you have already generated parameters (AMBER prepin and frcmod files) for any novel ligand/molecule that is present in your system. Info on how to do this can be found linked from the AMBER page.
Right! Lets begin:
Step 1: Preparing your pdb file
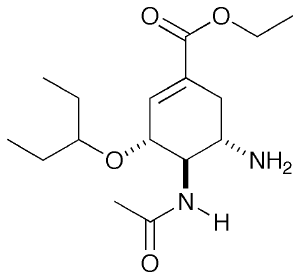
For this tutorial, I will be looking at neuraminidase (NA), an Influenza virus surface protein whose natural substrate is sialic acid. There are a few drugs which target this molecule as competitive inhibitors and one well known example is Tamiflu. Shown on the right is in fact the pro-drug form which you take. In the body, the ester is hydrolysed to a carboxylic acid and this is the form I will be looking at.
The starting point is a crystal structure of Tamiflu (oseltamivir) bound to NA which I obtained from the Protein Data Bank. NA is actually a tetramer so to cut down the simulation time, I will only consider the monomer and therefore I removed all but one copy from the PDB file, just by deleting the entries for the atoms.
Starting point: 2hu4.pdb
Removing Hydrogens from the protein
As AMBER uses an all-atom representation, we need to allow it to add in Hs as it sees fit when you generate the topology file. This is a problem if you already have Hs in the structure so for any part which you are going to load parameters for from the AMBER libraries, you must remove all H (including polar Hs) from the protein. This is best done using a script or a series of piped commands. For example, first, you find which types of H you have in the molecule using:
grep H 2hu4.pdb | more
You'll get something like this:
ATOM 2 H1 VAL 1 43.584 25.333 13.539 1.00 0.00 ATOM 3 H2 VAL 1 44.124 26.003 14.929 1.00 0.00 ATOM 4 H3 VAL 1 42.554 25.613 14.779 1.00 0.00 ATOM 12 H LYS 2 43.614 29.813 13.829 1.00 0.00 ATOM 19 HZ1 LYS 2 48.594 30.403 12.569 1.00 0.00 ATOM 20 HZ2 LYS 2 47.324 29.383 12.669 1.00 0.00 ATOM 21 HZ3 LYS 2 47.194 30.783 11.829 1.00 0.00 ATOM 25 H LEU 3 41.774 31.293 17.769 1.00 0.00 ATOM 34 H ALA 4 38.304 32.193 15.399 1.00 0.00 ATOM 40 H GLY 5 37.974 36.783 16.069 1.00 0.00 ATOM 45 H ASN 6 35.304 36.683 15.349 1.00 0.00 ATOM 51 HD21 ASN 6 34.094 36.233 14.939 1.00 0.00 ATOM 52 HD22 ASN 6 33.744 35.023 13.739 1.00 0.00 ...
The task now is to find a pattern which will let you remove each type of H in turn using grep -v. i.e. I can remove the first three types of H using:
grep -v ' H[1-3]' 2hu4.pdb
NOTE: I've got a space before the H there. This is so that I don't accidentally remove lines with NH1 on for example. A good method is just to do a normal grep for the pattern that you plan on using to check that you won't catch any atoms you don't intend to! Once you've found a patten that covers all types of H in your molecule, you need to string them together and redirect the output to a new pdb file e.g.
grep -v ' H[1-3] ' 2hu4.pdb | grep -v H[Z,D,H,G] | grep -v ' HE' | grep -v ' H ' > 2hu4_noH.pdb
The results is here: 2hu4_noH.pdb
Don't worry about the atom numbering being messed up, we'll deal with that later! Remember also that command presented above fits to 2hu4_noH.pdb file. Other PDB file can contain Hs with different names.
Adding H to the ligand
In my pdb file, there is a residue OTV corresponding to the drug molecule. When I generated parameters in AMBER for that residue, it had a complete set of Hs, and crystal structure from the PDB doesn't have any more. This is a problem! We will need to add them back in using e.g. Molfacture, a tool in VMD. You will need the latest version of VMD to do this so either download and compile it yourself (which is pretty easy - honest!), or use my binary located at
clust:~csw34/bin/vmd
Using Molfacture to do this is explained in a separate tutorial (as it's a bit long winded) in the VMD section. Once you have added all the Hs required and ensured that the charge on the molecule is what you require (-1 in this case due to a COO- group), you need to save the ligand you are editing as a new PDB using File>Save and changing the type to .pdb. The result is linked below:
Ligand plus Hs from VMD: vmdligand.pdb
NOTE: The coordinates have been preserved from the original crystal structure position of the ligand. This will turn out to be very useful later on!
Checking for consistent atom naming
This is a vital step as when you parametrised your ligand originally, each atom was assigned a name from the generalised forcefield. Unless the atoms names in your pdb match with those in the AMBER prepin file, you're not going to be able to make a topology or coordinate file to use with AMBER. Unfortunately, this just requires some manual editing. Here is one suggested method but feel free to change it if you think there is a more logical way forward. You will need a working copy of xleap for this bit. It is part of the AMBER package so if you do NOT have AMBER installed on your workstation, get someone to help you!
Here are the .prepin and .frcmod files for the OTV ligand that I made earlier (This is sounding a bit like Blue Peter!):
- prepin file: OTV.prepin
- frcmod file: OTV.frcmod
The first thing we need to do is create a .pdb from the .prepin file using xleap. Start xleap and type the following:
loadamberprep OTV.prepin savepdb OTV OTV.pdb quit
You will now have OTV.pdb in the directory. Looking at is, there is obviously something a bit wrong! Some of the atom names seem to start with numbers! LEaP is a very powerful tool (apparently), but as such - it has so many options and quirks that using it rapidly becomes a bit of a nightmare. In this case, it has messed around with the atom names and so we will need to correct this mistake. Open up OTV.pdb and OTV.prepin in your favourite editor and, atom by atom, ensure that the pdb file has the same atom names as in the prepin. For example, 1HM2 in the pdb file should clearly be HM21 and so on.
WARNING: DO NOT CHANGE THE PREPIN FILE AT ALL! Doing so will cause all sorts of bad things to happen. Only edit the pdb!
The resulting pdb file will look like this: OTV_checked.pdb. When you used Molfacture to add the Hs onto the ligand in the crystal structure, you also assigned them names but unfortunately, they will NOT be the same as those assigned by xleap (i.e. those we just checked from the prepi file) so now we need to compare OTV_checked.pdb with vmdligand.pdb visually. The best option is to open both pdb files in e.g. Pymol and orient them so they look the same, displaying the atom names from the label menu (aligning would be even better but in this case order of atoms in both PDB files is unfortunately different). Now, open vmdligand.pdb and, one atom at a time, check it's name is the same in both structures. If they are not, correct vmdligand.pdb with the right name.
The result is this: vmdligand_correct.pdb
Now, vmdligand_correct.pdb contains not only the correct atom names in the right order but also the correct xyz coordinates from the original crystal structure. We're ready to splice them back into the original pdb!
Creating the input pdb
Open the crystal structure you removed the Hs from earlier ( 2hu4_noH.pdb) and delete all the atoms belonging to the drug molecule. In their place, paste in the atoms directly from vmdligand_correct.pdb! Ensure that you add a TER line between the end of the protein and the start of the drug, and also between the end of the drug and the CA (Calcium) ion. We're now ready to have a go at making the topology file!
At this stage, we're working with: 2hu4_noH_corrected.pdb
Step 2: Making the initial topology file
Ok so now we have all the files required to generate a topology file using xleap:
- OTV.prepin - the prepin file for the ligand (that is not in the AMBER library)
- OTV.frcmod - the forcefield info file for the ligand
- 2hu4_noH_corrected.pdb - a pdb file with the same atom naming for the ligand as in the prepin file
We now use a simple script to construct the topology and coordinate files with xleap:
source leaprc.ff03 source leaprc.gaff loadamberprep OTV.prepin loadamberparams OTV.frcmod mol=loadpdb 2hu4_noH_corrected.pdb saveamberparm mol 2hu4.top 2hu4.cord savepdb mol 2hu4_leap.pdb quit
Save this in a file (say leap.in) and then run the following:
xleap < leap.in
You'll see a window open and lots of info scroll past. If you're quick enough you might even notice that we have a few problems with the input. This is almost bound to happen but luckily, xleap is pretty good at telling you how to fix them. So! Open up the leap.log file that has been generated and scroll through. After a bit of preamble (at line 841), you'll see the problems start i.e.
... Joining SER - LEU Joining LEU - CYS Joining CYS - PRO Joining PRO - ILE Created a new atom named: CD within residue: .R<ILE 12> Added missing heavy atom: .R<ILE 12>.A<CD1 14> Joining ILE - ASN Joining ASN - GLY Joining GLY - TRP Joining TRP - ALA ...
This message is repeated throughout the log file. What it means is that due to the definition of the ILE residue in the parameter libraries, AMBER is expecting the CD atom in ILE to be called CD1, not CD. We'll need to correct this and try running again. Before we do that though, there are other problems to address on line 1271:
... Created a new atom named: CD within residue: .R<CILE 385> Created a new atom named: O1 within residue: .R<CILE 385> Created a new atom named: O2 within residue: .R<CILE 385> Added missing heavy atom: .R<CILE 385>.A<O 19> Added missing heavy atom: .R<CILE 385>.A<OXT 20> Added missing heavy atom: .R<CILE 385>.A<CD1 14> Creating new UNIT for residue: CA sequence: 387 Created a new atom named: CA within residue: .R<CA 387> total atoms in file: 3007 ...
There are two problems here:
- The terminal ILE residue not only has the CD->CD1 naming issue but also the oxygens should be names O and OXT instead of O1 and O2.
- AMBER complains that it doesn't recognise the atom type CA which is the Calcium atom. Unfortunately there just aren't parameters in AMBER for Calcium so we need to either remove this line or change it to an ion for which the parameters exist e.g. Mg2+. This is what I will do in this case by changing the atom CA->MG in the pdb file (and the residue name from CA->MG2). I was able to do this by consulting the libraries for AMBER and noting which ions are parametrised. You can find the files in:
$AMBERHOME/dat/leap/lib
The file for ions is called ions94.lib. Note that the environmental variable $AMBERHOME is set when you install AMBER on your terminal. So to summarise, I need to make some changed to 2hu4_noH_corrected.pdb before AMBER will be happy to read it in, namely for all ILE residues I need to change the CD atom to CD1 and for the C terminal residue, O1->O and O2->OXT. Finally I need to change the Calcium ion (CA) to Magnesium (MG). This can be done manually or by using a find and replace script for say sed i.e. for the CD->CD1 change:
sed 's/ CD ILE / CD1 ILE /' 2hu4_noH_corrected.pdb > 2hu4_noH_final.pdb
Once all these edits are done - the leap.in script we used earlier should now work provided we change the filename. The final edited pdb file is called 2hu4_noH_final.pdb. Remove the leap.log file and then run the input again. Check the new log file for any problems before continuing. You often don't need to worry about the warnings concerning improper dihedrals because AMBER checks all possible impropers that could be defined, but many are not required and therefore there is no parameters for all of them. They can be important if you are trying to e.g. force planarity. It is worth checking the total charge is also what you expect. You should now have a few new files:
- 2hu4.top - the AMBER topology file
- 2hu4.cord - the input coordinate file
- 2hu4_leap.pdb - the corresponding final pbd file
Tweaking the topology file
Now we have an AMBER topology file but there are a few changes we need to make before we can use it. The first is necessary and the second, while not essential - is highly recommended. The first step involves renaming the terminal residues.
When xleap created the topology file, it also renames the terminal residues, something we didn't want it to do! Open up 2hu4.top in an editor and look for the section that begins with:
%FLAG RESIDUE_LABEL %FORMAT(20a4)
This is where the sequence is defined, however the N and C terminal residues should be named differently i.e. in our case VAL should be NVAL and ILE should be CILE. This edit isn't quite that simple however as the topology file uses a fixed format with four spaces per residue so you need to remove a blank space when you edit the name. e.g.
Originally we have:
... %FLAG RESIDUE_LABEL %FORMAT(20a4) VAL LYS LEU ALA GLY ASN SER SER LEU CYS PRO ILE ASN GLY TRP ALA VAL TYR SER LYS ...
And this should be changed to:
... %FLAG RESIDUE_LABEL %FORMAT(20a4) NVALLYS LEU ALA GLY ASN SER SER LEU CYS PRO ILE ASN GLY TRP ALA VAL TYR SER LYS ...
i.e. VAL_->NVAL where '_' is a space. The same should be done for the C terminal ILE residue. The resulting topology file is here:
Once you have make the changes, you need to run the script perm-prmtop.py as described on its page to ensure that the AMBER potential will be symmetric with respect to exchange of equivalent atoms, i.e.
./perm-prmtop.py 2hu4_fixed.top 2hu4_final.top
This script is still under development, please contact Edyta (em427) before using it!
You now have the final version of the topology file, 2hu4_final.top!
Step 3: Making a perm.allow file
If you are using OPTIM (with or without PATHSAMPLE), there is only one thing left to do before we are able to start running simulations with our system, and that is to create a perm.allow file. This is done using the perm-pdb.py script. Before we do this, we need to edit the 2hu4_leap.pdb as xleap has changed the terminal residue names here too, just like in the topology file. Open up 2hu4_leap.pdb and change the first residue, VAL->NVAL and the final residue ILE->CILE. This can be accomplished quickly using sed:
sed 's/VAL 1/NVAL 1/' 2hu4_leap.pdb | sed 's/ILE 385/CILE 385/' > 2hu4_leap_fixed.pdb
This time, the atom names being a bit funny (numbers first) doesn't matter! This gives you 2hu4_leap_fixed.pdb. Now copy perm-pdb.py into a file and run it as follows:
chmod 755 perm-pdb.py ./perm-pdb.py 2hu4_leap_fixed.pdb
This should produce a perm.allow file. You will also get a message asking you to check residues MG2 and OTV which is of course understandable as they are not simple amino or nucleic acids!
DONE! :)
Summary
So! The files which you will use in your AMBER simulations are:
- 2hu4_final.top (the topology file, usually called coords.prmtop)
- 2hu4.cord (the coordinate file, usually called coords.inpcrd)
- perm.allow - if you're using OPTIM or PATHSAMPLE
I hope that's helpful! Any errors or questions, feel free to contact me.
--csw34 16:42, 13 May 2008 (BST)