Preparing an AMBER topology file for a protein plus ligand system
When you want to use AMBER, either on its own to run MD, or interfaced with any of the group software (GMIN or OPTIM), you need to have two files at your disposal. These are the topology file (which describes the connectivity of atoms, angles etc) and the coordinate file (which shockingly defines the coordinates of the atoms in your system).
For big systems, generating these files can take a bit of time, and the difficulty increases when you are trying to simulate something for which parameters do not exist in the AMBER libraries i.e. non-standard amino acids, or, more usually, a system with a novel ligand (for example a drug molecule). This tutorial will deal with a big system, hopefully covering all bases. You will probably find the process much less arduous when you are dealing with something smaller and I will try to point out where I am deviating from what you would need to do if you only had say a protein composed of standard amino acids.
I will also assume that you have already generated parameters (AMBER prepin and frcmod files) for any novel ligand/molecule that is present in your system. Info on how to do this can be found linked from the AMBER page.
Right! Lets begin:
Step 1: Preparing your pdb file
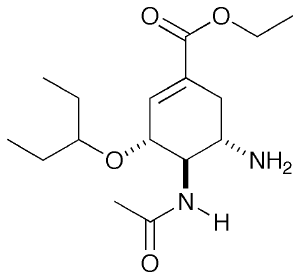
For this tutorial, I will be looking at neuraminidase (NA), an Influenza virus surface protein whose natural substrate is sialic acid. There are a few drugs which target this molecule as competitive inhibitors and one well known example is Tamiflu. Shown on the right is in fact the pro-drug form which you take. In the body, the ester is hydrolysed to a carboxylic acid and this is the form I will be looking at.
The starting point is a crystal structure of Tamiflu (oseltamivir) bound to NA which I obtained from the Protein Data Bank. NA is actually a tetramer so to cut down the simulation time, I will only consider the monomer and therefore I removed all but one copy from the PDB file, just by deleting the entries for the atoms.
Starting point: 2hu4.pdb
(a) Removing Hs from the protein As AMBER uses an all-atop representation, we need to allow it to add in Hs as it sees fit when you generate the topology file. This is a problem if you already have Hs in the structure so for any part which you are going to load parameters for from the AMBER libraries, you must remove ALL H. This is best done using a script or a series of piped commands. For example, first, you find which types of H you have in the molecule using:
grep H 2hu4.pdb