Using VMD to display and manipulate '.pdb' files
VMD's default input format is pdb and so it isn't necessary to specify a file extension if loading from the command line, so the following syntax could be used to load several pdbs:
vmd -f pdb1.pdb -f pdb2.pdb -f pdb3.pdb
Using Molefacture
Once the PDB is loaded, it is possible to edit it using the Molefacture plug-in from the Extension > Modelling menu (the selection syntax is the same as for generating graphical representations). N.B. Molefacture doesn't like molecules with hundreds or thousands of atoms, nor is it easy to manually edit them, it might be easier to load a smaller portion of the molecule into Molefacture (e.g. a particular residue you want to edit in a protein) and then paste the output pdb into the original pdb of the larger molecule.
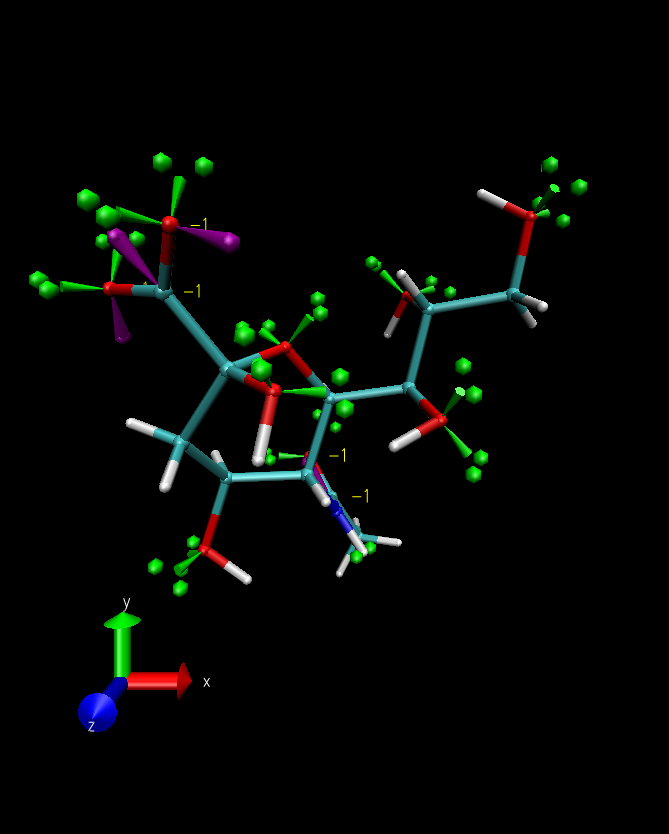
Once you have loaded your selection into Molefacture, it will be displayed in the VMD display window with lone pairs highlighted as green lobes with two spheres and empty orbitals displayed as purple lobes. Charged atoms will also be labelled with their net charge. These can be switched on and off in the Settings menu of the Molecule Builder window.
Atoms are selected by clicking on them and this allows you to use the Atoms options in the Molecule Builder window. These can add hydrogen, delete the atom, alter its geometry and oxidation state and finally edit the atom. Editing the atom allows you to manually specify its name (for the pdb you generate), its type, partial charge and element.
To select multiple atoms, hold shift whilst clicking. If two atoms are selected, the Bonds options can be used. This allows you to adjust the bond length, bond order and the bond dihedral (where the other two groups defining the dihedral are highlighted automatically by Molefacture in yellow).
If three atoms are selected, the angle option becomes available and can be used to specify the bond angles between two groups bonded (Atom1 and Atom3 in the list) to a central atom (Atom2). It is also possible to select whether you wish to move Group1 (attached to Atom1), Group2 (attached to Atom3) or both with the relevant tickboxes (default is both). If you wish to reset the geometry of Atom2 to a standard (e.g. tetrahedral or planar) you can alter this again with the relevant Atoms options.
Finally, the Build menu has functionality to automatically type bonds and add hydrogens to all atoms to neutralise the molecule (not recommended). It is also possible to replace selected hydrogens with a fragment or to clear the molecule and start from a predefined fragment.
Those for replacing hydrogens are:
- Phenyl
- Thiol
- Carboxyl
- Butyl
- Amino
- T-butyl
- Formyl
- Vinyl
- Chloride
- Hydroxyl
- Methyl
Those you can start a new molecule from are:
- Naphthalene
- Cyclobutane
- Tetrahydrofuran
- Water
- Isoprene
- Cyclopentane
- Cyclopropane
- Acetone
- Ammonia
- Methane
- Cyclohexane
- Benzene
- Butane
There is also a protein builder...if anyone knows how to use this properly...
Molefacture also creates a file called Molefacture_tmpmol.xbgf in your current directory which seems to be intended as a temporary file, but isn't removed when Molefacture ceases running, so you may want to remove this after use to stop it cluttering your directory.