Producing sexy ray-traced images
Pardon the title but you'll understand when you see the results!
Before you ray trace
Before you start playing with ray tracing, you need to be sure that you're happy with the way your system is represented. There are LOADS of options in Pymol to change how things appear, and most of them are not available in the GUI. For a complete list, go to Settings>Edit All and have a play around!
So, you've got a PDB, loaded it into Pymol and arranged it so that it looks how you'd like - some bits as a cartoon, some as sticks etc. How do you go about making an image that you can then use for a poster, or a paper, or a CPGS/thesis? The answer lies in ray tracing. To get Pymol to actually produce a ray traced image is remarkably easy, just type
ray
in either the viewing window, or the GUI and press enter. After a few seconds (progress is indicated by a bar at the top of the viewing window), you will see the image suddenly change! You could now save this image using
png raytraced.png
BUT you'd quickly find that
- the image quality isn't up to much and so you can't use it in a publication
- it's too small to be used on a poster
- a black background is NEVER a good idea for images you might print for obvious reasons
To resolve these issues, I will now go through the key settings you should ALWAYS have set,
Recommended settings
set ray_trace_fog,0 set ray_shadows,0 unset depth_cue bg_color white set antialias,2 set hash_max, 300
You may actually want to have shadows in your image, but for anything larger than a small peptide, it can become very distracting. If you type ray again now, you can already see the improvement. 'hash_max' is very important as it defines the maximum amount of memory Pymol can use while ray tracing and defaults to a wimpy 100mb. Setting it to 300 seems to be around the point where further increases give no return. Make sure you have enough RAM though! But...there is more to come! Pymol supports a selection of ray tracing modes, four in fact. Which one you choose to use depends largely on which effect you require. Modes are selected with the command:
set ray_trace_mode, X
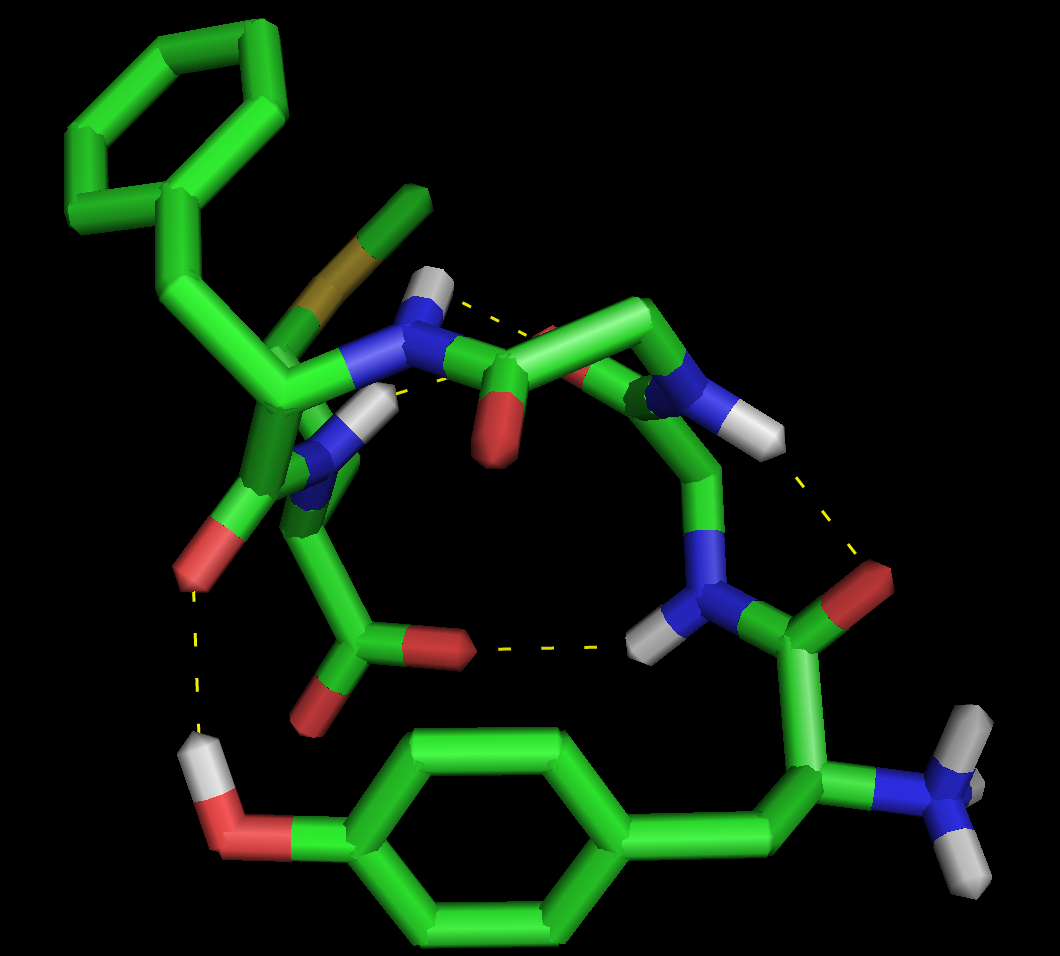
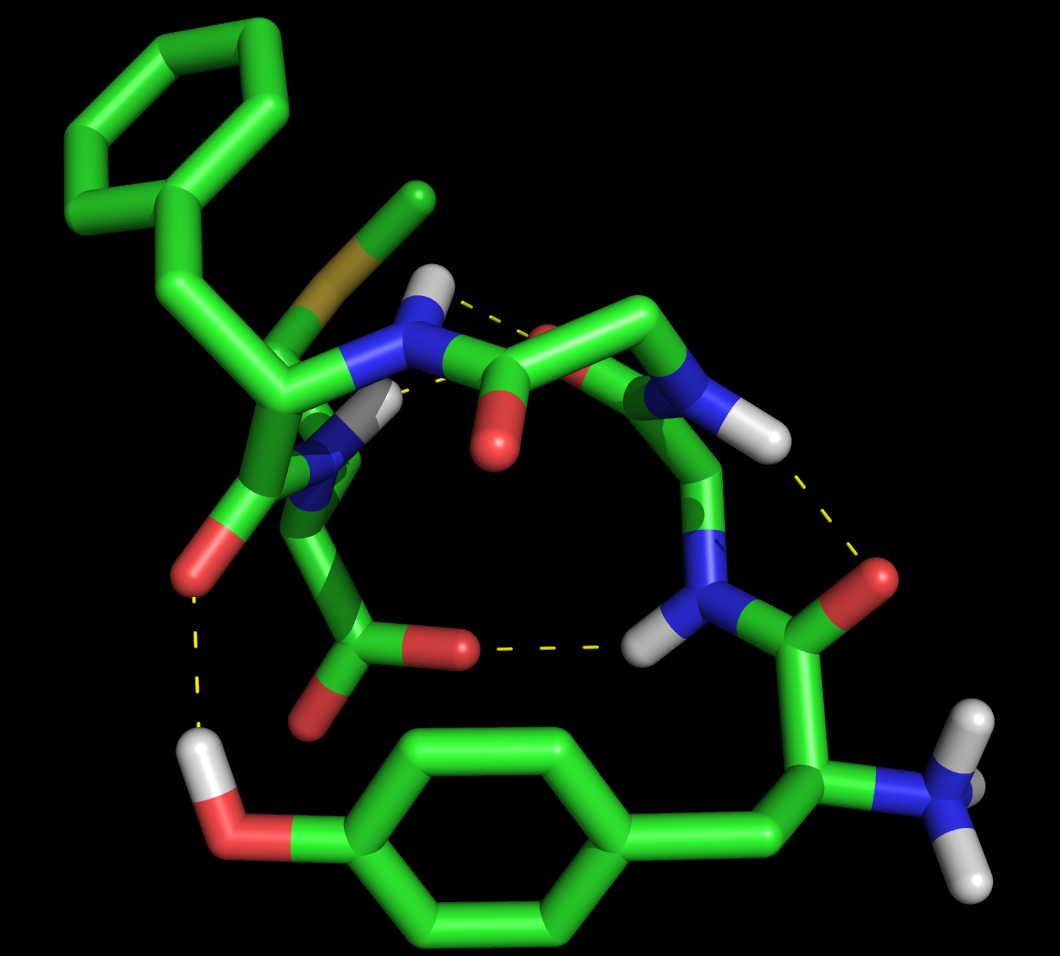
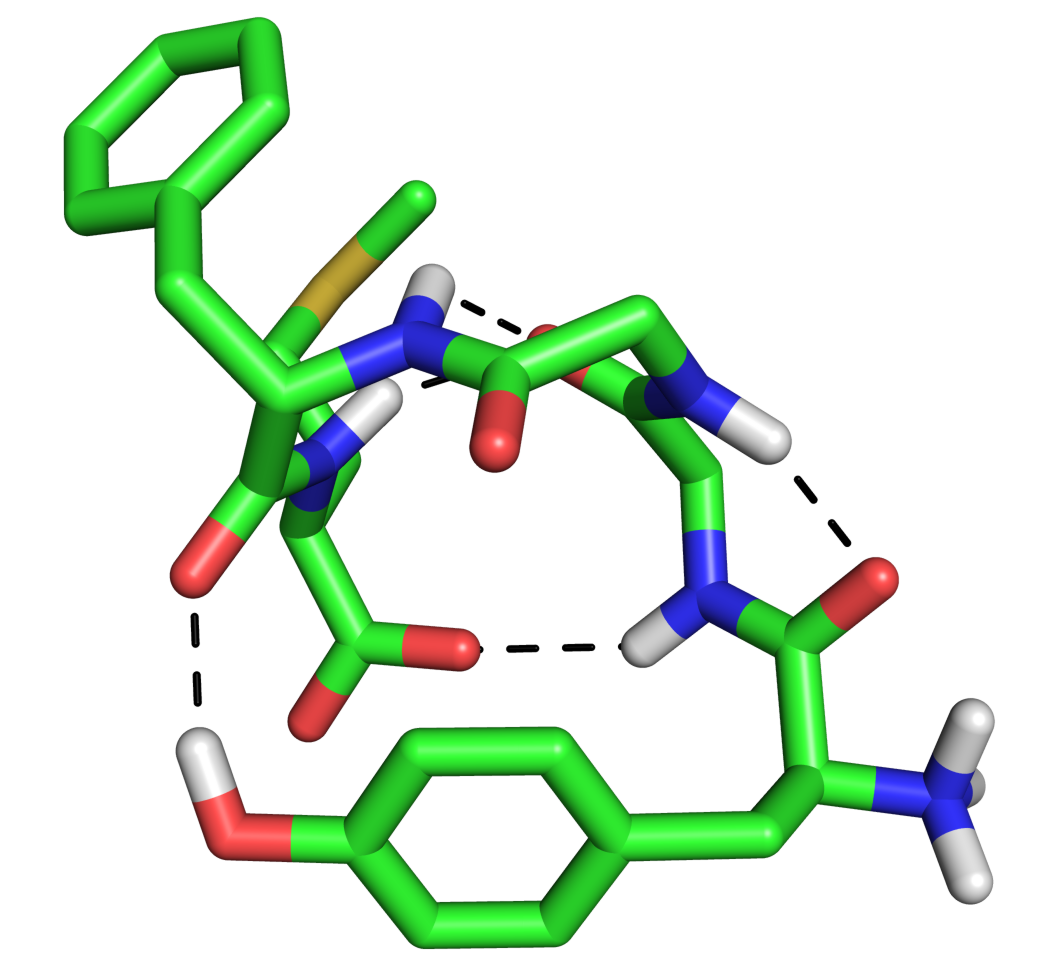
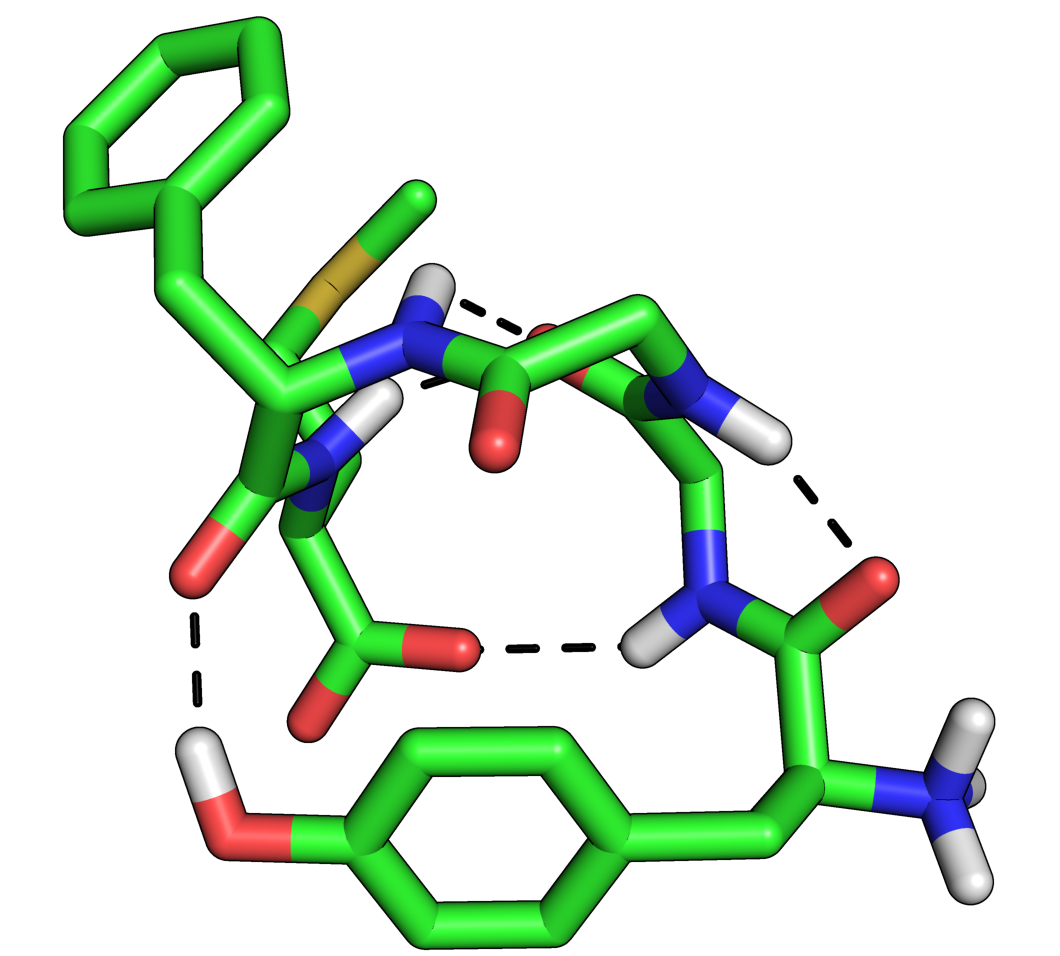
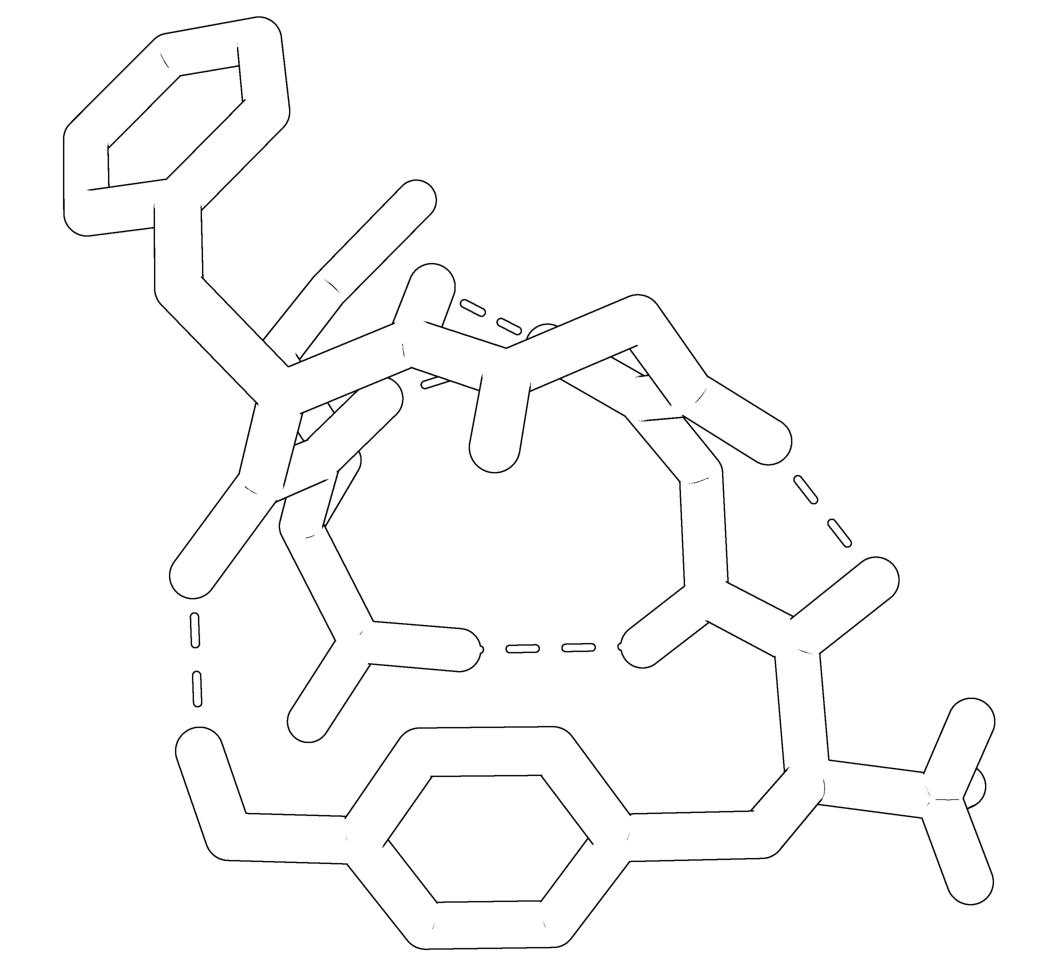
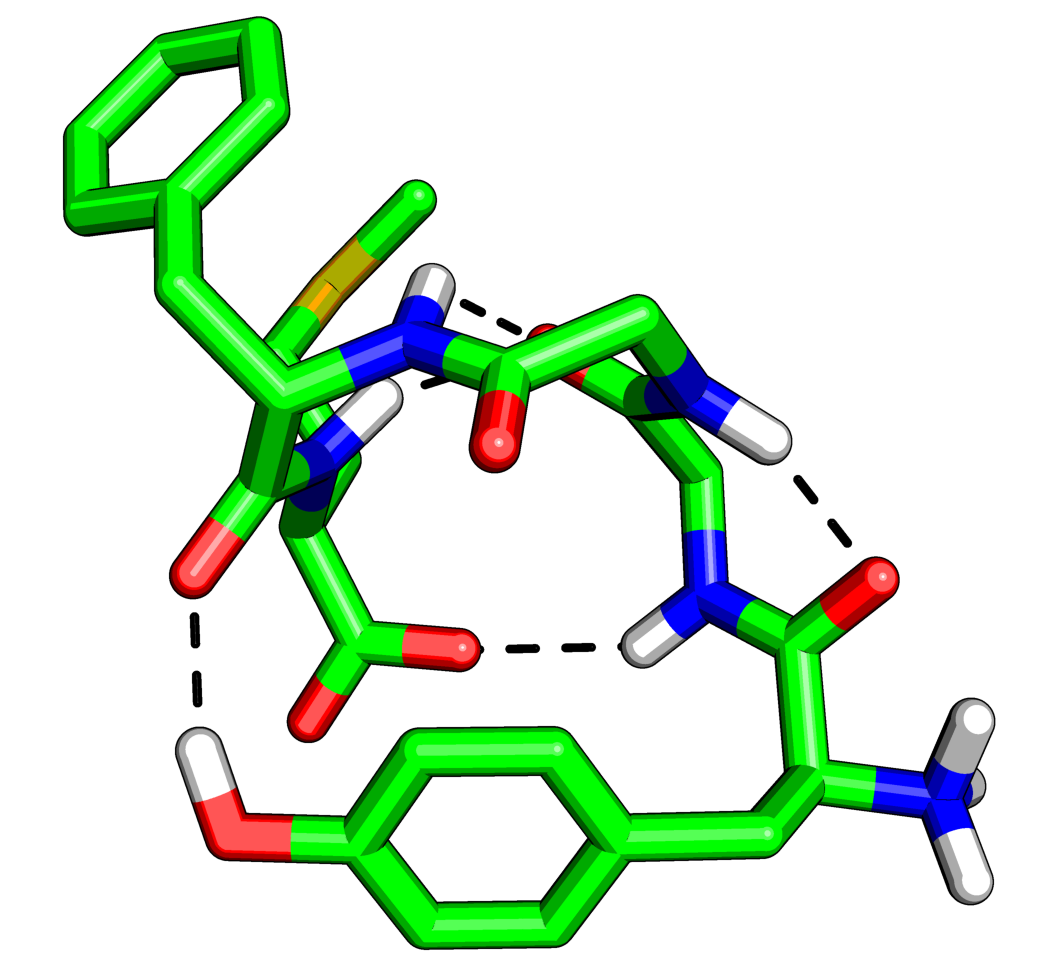
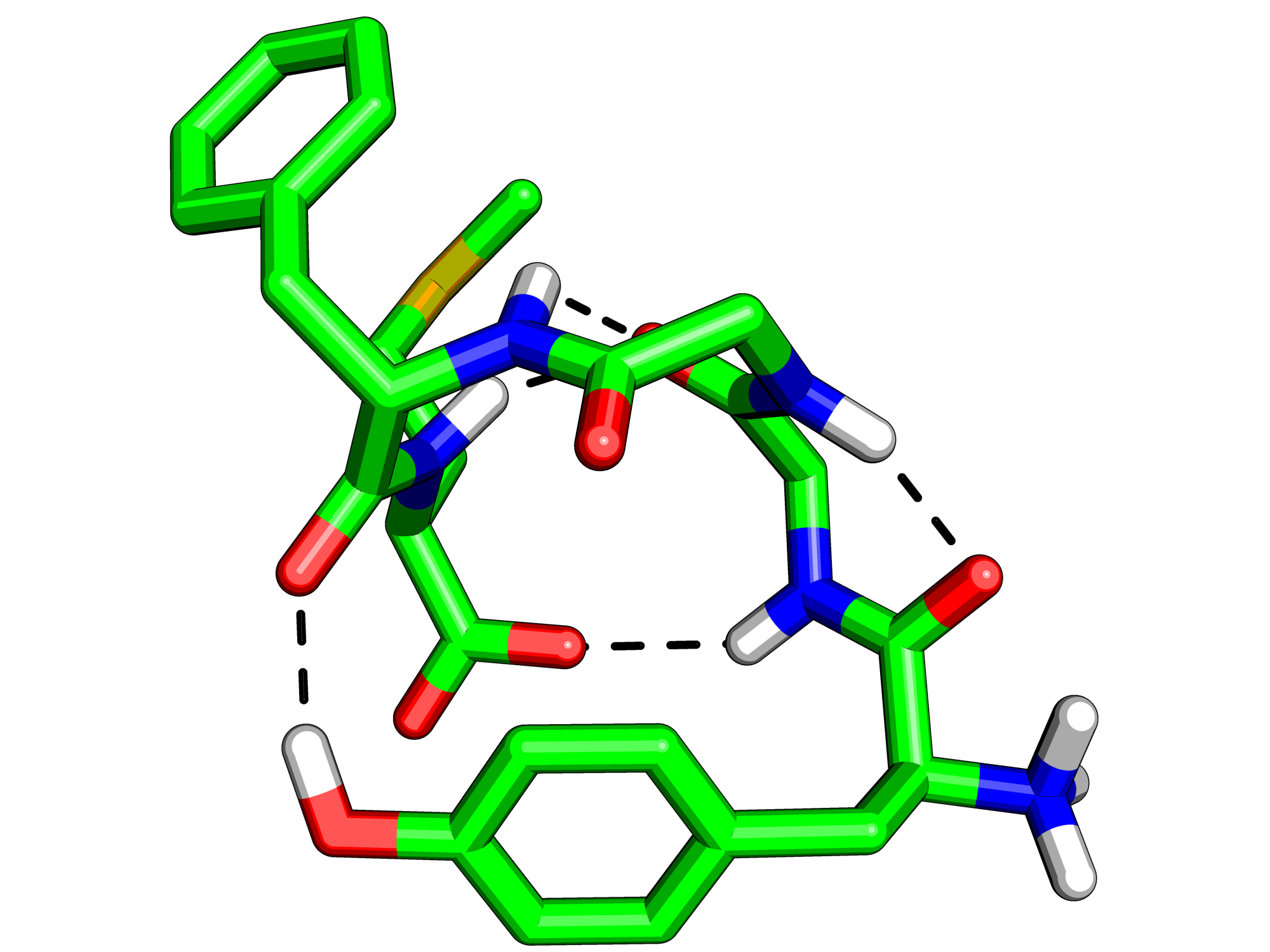
where X is 0,1,2 or 3. 0 is the default, 1 puts a black outline around the molecule, 2 gives you a black and white 'sketch' and 3 gives you a funky cartoon like image, with quantised colouring! All four are demonstrated in the images here. My personal favourite is mode 3 although you should bear in mind that it might not look as good when printed.
Making images for publication
Once you've chosen your ray tracing options, you're ready to produce the final image. As you'll likely want it to be publication quality, I recommended using the following
ray 2000,1500 png raytraced.png, dpi=300
These settings can of course be changed. You might find that you need to change the resolution if you have a wide-screen monitor (2000x1500 is for 4:3 aspect ratio monitors) as you might end up inadvertently chopping bits off the image. If in doubt, you can always just use the resolution your desktop is set to. PyMOL's "ray X, Y" command means that a ray-traced image of size X-by-Y dots will be produced. So if you want a 300 dpi, 10" x 6" image, then you'd do "ray 3000,1800"
Voila! you are now the proud owner of a rather good looking image! For more info, you should check out the Pymol Wiki.
Once you have a pretty png file you may need it in a compressed tiff format for a journal. This is easily done via the convert program which is a member of the ImageMagick suite. The compression is sometimes necessary.
convert -compress lzw raytraced.png raytraced.tiff