CamCASP/Notes/3
CamCASP => Notes => Energy scans for display
Energy scans for Display using ORIENT
Obtaining the grid
ORIENT calculates the surface around the molecule. So we need to setup an ORIENT file to obtain this grid of points. This grid will then be passed to CamCASP which will calculate the interaction energies (next step).
We will use the CLUSTER program to help construct the ORIENT command file.
Title Pyridine : properties cluster file Global CamCASP /home/ajm/SITUS/current Units Bohr Degree Overwrite Yes End Molecule pyridine ! Optimzed with PBE0/cc-pVTZ Gaussian03 ! C2v symmetry Units Angstrom H1 1.0 -2.050322 1.274414 0.000000 H2 1.0 -2.147113 -1.203259 0.000000 H3 1.0 0.000000 -2.487558 0.000000 H4 1.0 2.147113 -1.203259 0.000000 H5 1.0 2.050322 1.274414 0.000000 N 7.0 0.000000 1.382844 0.000000 C1 6.0 -1.134410 0.690452 0.000000 C2 6.0 -1.190513 -0.695795 0.000000 C3 6.0 0.000000 -1.403912 0.000000 C4 6.0 1.190513 -0.695795 0.000000 C5 6.0 1.134410 0.690452 0.000000 End Files Molecule pyridine Basis daTZ File-prefix pyridine Orient files for display Interface file Memory 2000 MB End Finish
CLUSTER will create three file. The *.template file is not needed. You can delete it.
Copy pyridine_display.ornt to pyridine_display_grid.ornt. Edit pyridine_display_grid.ornt so that:
1. The radius of any polar hydrogen (one that can participate in an hydrogen-bond) is set to zero. You can do this using
Types Hp Z 1 Radius 0.0 ... End
at the start of the file. And in the Molecule block set all polar hydrogens to Type Hp. For more details please see the ORIENT manual. By the way, you do not need to do this, but doing so makes the scan more true to reality as the distance of close-approach to such hydrogen atoms is smaller than that for other hydrogen atoms.
2. Comment out the statements in the Polarizabilities block. We will not need polarizabilities to construct the grid.
3. Un-comment the Write command in the Display energy block.
Now run ORIENT
orient < pyridine_display_grid.ornt
You will see a blob around the molecule. This is OK. Make sure the blob looks roughly like you think it would. There will be no colours. This too is OK. Also, ORIENT will have produced the file pyridine_2vdW.grid that contains the grid points and triangles. We need only the grid points. So copy pyridine_2vdW.grid to pyridine_2vdW_points.grid and this file as follows:
1. Comment out the first line which contains the number of points in the grid and
2. delete the list of triangles.
Here's what it should look like:
$ more pyridine_2vdW_points.grid
# 2370 4736
-9.00000000 -1.50000000 -0.63302610
-9.00000000 -2.18117397 0.00000000
-9.03969412 -1.50000000 0.00000000
...
... 2370 sets of coordinates...
NOTE: By default the surface constructed is the so-called 2 x vdW surface. Basically, double the van der Waals radii of the atoms (except polar hydrogens), stretch a rubber sheet over those spheres, construct the grid of points on the rubber sheet. Well, that's it conceptually. The ORIENT manual has the details. What's important is to realise that you can change the parameters of this surface. For example, you may want a 1.8 x vdW surface. Have a peek in the Display energy block and tweak the parameters.
CamCASP energy scan
Now we need to use this list of points to perform a CamCASP energy scan. Only this scan will be with the molecule (pyridine) and a spherical probe (a neon atom for the dispersion and a +1 charge for the electrostatics and induction). This is not necessary, but is very convenient.
The CamCASP file pyridine.cks that has been produced by CLUSTER is not in the correct format. We need to add the description of the probe molecule (neon) and the commands for the energy scan. You will also need to copy the MO and Hessian files for the molecule (pyridine) and neon atom to this directory. I have assumed you have these at hand. If not, calculate them (I'll describe this later)
Copy pyridine.cks to pyridine_scan.cks and edit it to look like:
TITLE pyridine ... neon / Q energy scan
TITLE Basis d-aug-cc-pVTZ
MEMORY 2454 MB
SET Global_data
CamCASP-path /home/ajm/SITUS/current
Units Bohr cm-1
Scf-code Dalton
XC-func PBE0
Overwrite yes
END
MOLECULE pyridine at 0.0 0.0 0.0
Charge 0
Echo No
Hessian format SAPT2006
MO-file mo-pyridine.data
H1-file h1-pyridine.data
Basis Main
...
End
Basis Aux
...
End
END
Molecule Ne at 0.0 0.0 0.0
Charge 0
Echo No
Hessian format SAPT2006
MO-file mo-ne-daTZ.data
H1-file h1-ne-daTZ.data
Basis Main
Spherical
Units Bohr
Format GAMESS
Ne 10.0 0.00000000 0.00000000 0.00000000 TYPE Ne
#include-camcasp basis/gamess_us/d-aug-cc-pVTZ/Ne
---
End
Basis Aux
Cartesian
Units Bohr
Format TURBOMOLE
Ne 10.0 0.00000000 0.00000000 0.00000000 TYPE Ne
Limit G
#include-camcasp basis/aux/aug-cc-pVQZ/Ne
---
End
End
SET QUAD
Type Gauss-Legendre
Beta 0.5
END
BEGIN DF
Molecule pyridine
Type FULL
Eta = 0.0
Lambda = 0.0
Print only normalization constraints
END
BEGIN DF
Molecule Ne
Type FULL
Eta = 0.0
Lambda = 0.0
Print only normalization constraints
END
SET PROPAGATOR
Type CKS
DF without constraints
DF-integrals
END
Begin Energy-scan
Probe pyridine with Ne and Charge +1.0
Scan E2ind & E2disp & E1elst
Units Bohr
Points
Translations-only
#include pyridine_2vdW_points.grid
End
End
FINISH
And I have the MO and Hessian files:
ls h1-ne-daTZ.data pyridine_2vdW.grid pyridine_daTZ_DMA2_L4.mom h1-pyridine.data pyridine.axes pyridine_display_grid.ornt mo-ne-daTZ.data pyridine.cks pyridine_display.ornt mo-pyridine.data pyridine.clt
Now we can run CamCASP using
runcamcasp pyridine_scan -q intel.q --nochecks
If all went well, you should have the files
$ls OUT/ pyridine_scan_camcasp.out energy_file.dat
Preparing data files for ORIENT
The energies in energy_file.dat cannot be read by ORIENT. First of all, we need to decide on the kind of units suitable for the problem. With the CamCASP command file described above, the energies will be in cm-1. This is almost certainly not suitable for the electrostatic and polarization (induction) energies as these energies tend to be large. A more convenient unit for the energies may be kJ/mol. See Odds and Ends for instructions about making this conversion. We will assume that the energies are now in kJ/mol.
Now copy the grid file (see above) pyridine_2vdW.grid to pyridine_Q_elst_2vdW.grid, pyridine_Q_ind_2vdW.grid and pyridine_Ne_disp_2vdW.grid. Edit pyridine_Q_elst_2vdW.grid to include the electrostatic (E(1)elst) energies from energy_file.dat (in kJ/mol) alongside the grid points. Do the same for the induction and dispersion files. Here's what the dispersion file pyridine_Ne_disp_2vdW.grid should look like:
$ more pyridine_Ne_disp_2vdW.dat
2370 4736
-9.00000000 -1.50000000 -0.63302610 -0.185652E+01
-9.00000000 -2.18117397 0.00000000 -0.185379E+01
-9.03969412 -1.50000000 0.00000000 -0.184659E+01
-9.00000000 -1.50000000 0.63302610 -0.185652E+01
...
...
...
9.03969412 -1.50000000 0.00000000 -0.184659E+01
9.03304707 -0.75000000 0.00000000 -0.184386E+01
9.00455618 0.00000000 0.00000000 -0.184341E+01
1 2 3
3 2 4
1 5 6
...
...
...
The dispersion energies are in the fourth column. The integers at the bottom are the triangles used by ORIENT to construct the 3D image.
Orient command files for display
Now we are in a position to construct the ORIENT command files for the OpenGL display. We will display only energies here. To find out how to use the molecular moments, polarizabilities and dispersion coefficients to generate energies for a display or compare with the computed SAPT(DFT) energies, see The ORIENT display .
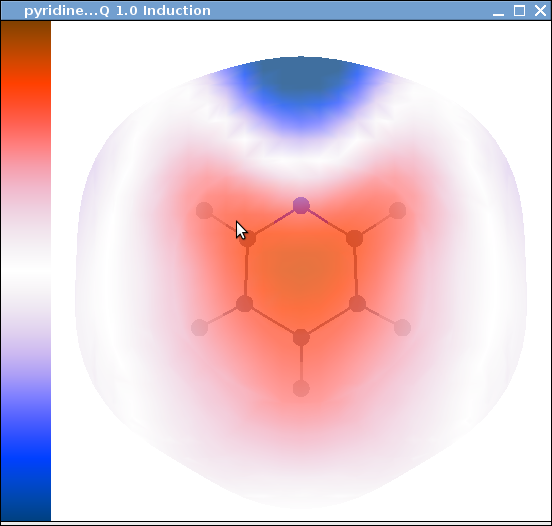
Our starting point will be the pyridine_display_grid.ornt file we have already created for the ORIENT grid calculation. Copy it to pyridine_ind.ornt. And edit the Display Energy section so that the file looks as follows:
$ more pyridine_ind.ornt
! ORIENT display commands
UNITS BOHR
Parameters
Sites 13 polarizable 12
S-functions 50000
Alphas 50000
Parameter-sets 50000
Pairs 100000
End
Types
H1 Z 1
H2 Z 1
H3 Z 1
H4 Z 1
H5 Z 1
N Z 7
C1 Z 6
C2 Z 6
C3 Z 6
C4 Z 6
C5 Z 6
End
Molecule pyridine at 0.0 0.0 0.0
! Units BOHR
H1 -3.87454677 2.40829326 0.00000000 Type H1
H2 -4.05745524 -2.27382980 0.00000000 Type H2
H3 0.00000000 -4.70080300 0.00000000 Type H3
H4 4.05745524 -2.27382980 0.00000000 Type H4
H5 3.87454677 2.40829326 0.00000000 Type H5
N 0.00000000 2.61319624 0.00000000 Type N
C1 -2.14372406 1.30476509 0.00000000 Type C1
C2 -2.24974336 -1.31486189 0.00000000 Type C2
C3 0.00000000 -2.65300899 0.00000000 Type C3
C4 2.24974336 -1.31486189 0.00000000 Type C4
C5 2.14372406 1.30476509 0.00000000 Type C5
! #include pyridine_DMA2_L4.mom
End
Edit pyridine
#include pyridine.axes
Bonds Auto
End
Polarizabilities for pyridine
! Assumed that the pols are in the local-axes
! So they are read after axes are defined
! Read rank <WSMLIMIT! 2>
! #include pyridine_ref_wt4_L<WSMLIMIT!2>_static.pol
! End
! Limit rank <WSMLIMIT! 2> for +++
! H1 H2 H3 H4 H5 N C1 C2 C3 C4 +++
! C5
End
Pairs
! Dispersion damping factor <BETA!2.0>
! Induction damping factor <BETA!2.0>
! #include <POTENTIAL!atom_atom>
End
Units Bohr kJ/mol
! This is a probe of the induction energy
Atom X at 0.0 0.0 0.0 rank 0
Q00 = 1.0
End
Display energy
Title "pyridine...Q 1.0 Induction "
Molecule pyridine
! * Use the Compare line for energy comparisons and
! * comment out the Radii, Step and Grid lines
! Compare with energy_values.dat
import file pyridine_Q_ind_2vdW.dat with values
! Radii scale 2.0
! Step 0.75 B
! Grid exp
Colour-map
0 210 0.25 1
6 240 0.75 1
12 300 1.0 0
18 360 0.75 1
24 390 0.25 1
End
Viewport 10
Colour-scale min -25 max -19 top -19
Probe X
Ball-and-stick
! * Uncomment the following line for the grid:
! Write pyridine_2vdW.grid no values
End
Finish
This is the entire file just so that you can see the entire thing at least once! And here is the axis file (see the ORIENT manual for details):
$ more pyridine.axes Axes N z from C3 to N x from N to C1 C1 z from C1 to H1 x from C1 to C2 H1 z from C1 to H1 x from C1 to C2 C5 z from C5 to H5 x from C5 to C4 H5 z from C5 to H5 x from C5 to C4 C2 z from C2 to H2 x from C2 to C3 H2 z from C2 to H2 x from C2 to C3 C4 z from C4 to H4 x from C4 to C3 H4 z from C4 to H4 x from C4 to C3 C3 z from C3 to H3 x from C3 to C4 H3 z from C3 to H3 x from C3 to C4 End
What have we done? There is a new line in the last section:
import file pyridine_Q_ind_2vdW.dat with values
This tells ORIENT to read in the grid from the file and also the values (energies). Since the grid is in this file, we don't need the commands that generate the grid, so we have commented out the lines:
! Radii scale 2.0 ! Step 0.75 B ! Grid exp
We also need to decide on an energy range for the display. This is normally done by trial and error. ORIENT prints out the energy range, so you can use this range in the line:
Colour-scale min -25 max -19 top -19
And if you now run ORIENT you should get the figure.