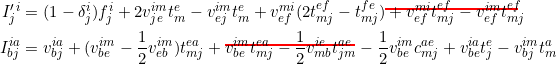GAMESS
Obtaining GAMESS
You can request for the latest version of GAMESS for free here.
Compilation
GAMESS has an unusual but straightforward system for compiling: untar the archive and run the ./configure script, and answer all the questions. For the most basic installation, choose socket for the parallelisation. The script will ask if you want to install a number of packages, and they are all completely optional.
After the configuration, do not follow machines/readme.unix but instead follow the instructions in ./README.md, i.e., run make ddi, make modules (if not using libxc), and then make -j${nproc}.
Running GAMESS
Set up running script
After a successful compilation and linking, you should see the script ./rungms appear in the GAMESS root directory. You need to edit this file, specifically the first four lines after the comments. You should create two directories, restart and userscr, and point the script to them, and also set GMSPATH to the actual installation location.
After all this, you can run GAMESS by invoking
rungms [input_file] 00 1 1 1
where 00 is the version number you'll have specified in the configuration stage, and the three 1's are to do with parallelisation options, here we run the most basic serial version.
Input file
You can find example input files under ./tests/, and the complete documentation here.
Geometry
The default geometry specification is by only specifying symmetrically unique atoms, but you can also use cartesian and Z matrix format. This is controlled by a $contrl group keyword.
Documentation on point group symmetry can be found by searching point group in the documentation. Briefly, C2v is cnv 2 and D_infh, despite their advice to use dnh 4, you should use dnh 2, otherwise results won't agree with other packages.
Consistency with other packages
GAMESS also freezes core electrons in post-HF methods by default, and this may lead to slightly different results compared to other programmes. To turn it off in coupled-cluster for example, add the ncore=0 keyword in the $CCINP group.
Basis sets
GAMESS doesn't use standard basis names, to find out the internal names, search $BASIS in the documentation above. Specifically, it doesn't have a lot of the def2 family of basis sets. You can paste the basis sets into the input file from Basis Set Exchange, but this requires that you use their own geometry specification standards (symmetrically unique atoms).
Sample input file =
Finally, an example input for a water molecule at 1.6 A bond length and 104.45 bond angle with a restricted HF reference at cc-pVDZ/CR-CCSD(T) level is given below:
$contrl scftyp=rhf coord=zmt runtyp=energy units=angs cctyp=cr-cc ispher=1 $end $system mwords=100 memddi=500 $end $guess guess=huckel $end $ccinp maxcc=100 ncore=0 $end $basis gbasis=ccd $end $data cnv 2 O H 1 1.6 H 1 1.6 2 104.45 $end
All keywords are case insensitive. Note the single whitespace in front of each keyword group, which seems to be important..
Issues with its coupled cluster modules
GAMESS implements a great variety of coupled cluster methods, including the renormalised and the completely renormalised CCSD(T) and [T]. The reference paper for all of these methods (CCSD, CCSD(T)/[T], (C)RCCSD(T)/[T]) is found here. Unfortunately if you ever tried to code these equations up from scratch yourself, you'll find that they're wrong. After extensive debugging, we found that there are two typos in Table 1 defining the intermediates needed in the CCSD amplitude update equations: