Difference between revisions of "The effect of calculating less than the maximum number of eigenvalues using ENDHESS n"
Jump to navigation
Jump to search
import>Csw34 |
import>Csw34 |
||
| Line 5: | Line 5: | ||
[[Image:Nendhessflu1merDSYEVR.png|Plot showing the effect of reducing the number of eigenvalues calculated for a large system, the 3557 atom HA 1mer for Indonesia H5 complexed with (2,3) linked sialic acid|center]] |
[[Image:Nendhessflu1merDSYEVR.png|Plot showing the effect of reducing the number of eigenvalues calculated for a large system, the 3557 atom HA 1mer for Indonesia H5 complexed with (2,3) linked sialic acid|center]] |
||
| − | Note that the time saved is rather small |
+ | Note that the time saved is rather small, with only just over a minutes difference between calculating ALL eigenvalues and NONE! |
Latest revision as of 14:19, 2 December 2009
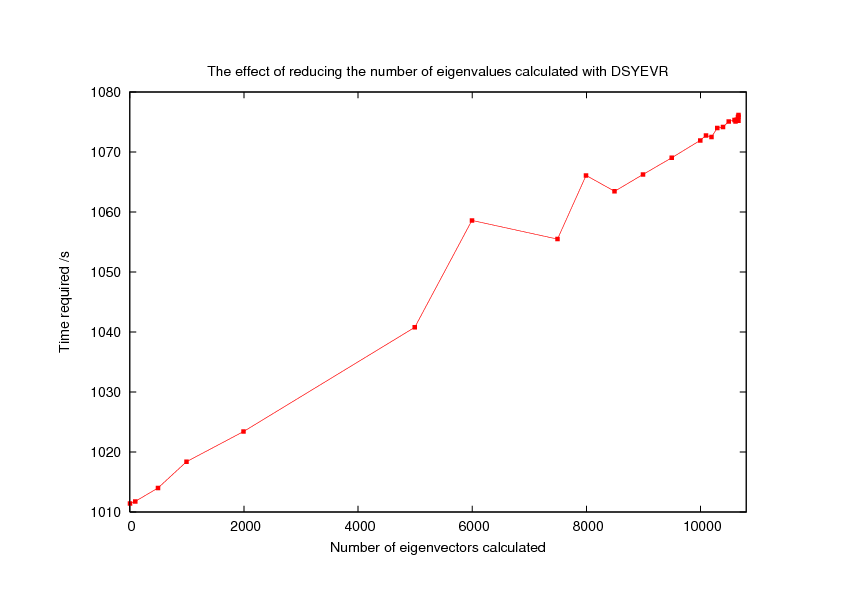
For large systems, calculating the eigenvalues (and eigenvectors) of the Hessian can be extremely time consuming. As a result, it may be helpful to calculate only a subset of eigenvectors. This is possible using an argument to the ENDHESS keyword. If you specify ENDHESS n where n is less than 3*NATOMS, the total number of eigenvalues.
Here is a plot showing the effect of reducing the number of eigenvalues calculated for a large system, the 3557 atom HA 1mer for Indonesia H5 complexed with (2,3) linked sialic acid:
Note that the time saved is rather small, with only just over a minutes difference between calculating ALL eigenvalues and NONE!