GMIN: Difference between revisions
Jump to navigation
Jump to search
import>Csw34 |
import>Mp466 |
||
| (3 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:Gmin.jpg|thumb|GMIN in all its glory|300px|right]] |
[[Image:Gmin.jpg|thumb|GMIN in all its glory|300px|right]] |
||
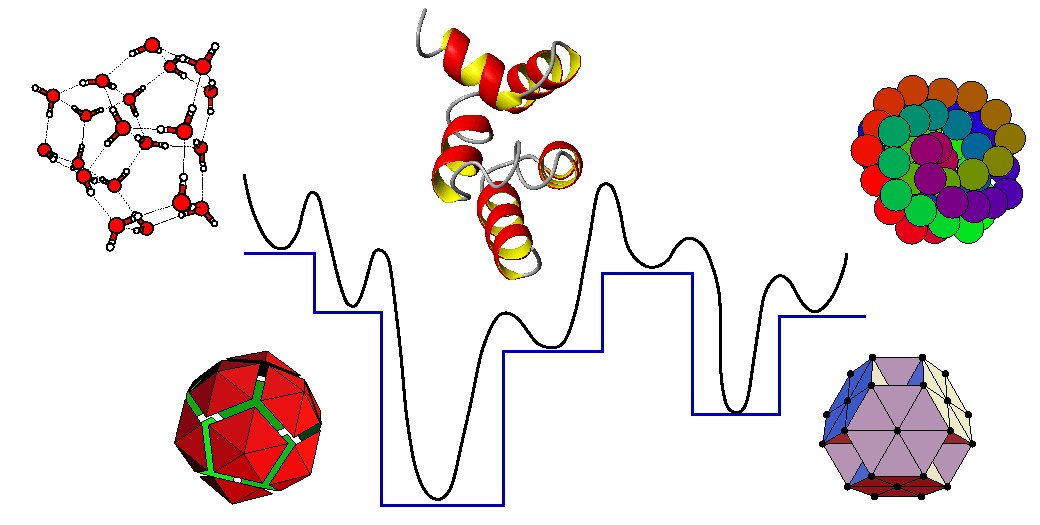
A program for finding global minima and calculating thermodynamic properties from basin-sampling. The GMIN homepage is located [http://www-wales.ch.cam.ac.uk/GMIN/ here]. |
A program for finding global minima and calculating thermodynamic properties from basin-sampling. The GMIN homepage is located [http://www-wales.ch.cam.ac.uk/GMIN/ here]. |
||
== Compiler compatibility == |
|||
*[[pgi64/8.0/1]] |
|||
== Tutorials == |
== Tutorials == |
||
| Line 12: | Line 15: | ||
* [[Optimization of ligand docking using AMBER9]] |
* [[Optimization of ligand docking using AMBER9]] |
||
* [[Restarting a GMIN run from a dump file]] |
* [[Restarting a GMIN run from a dump file]] |
||
* [[Using the implicit membrane model IMM1]] |
|||
* [[Running a Go model with the AMHGMIN]] |
|||
== Scripts == |
== Scripts == |
||
Latest revision as of 18:40, 26 July 2009
A program for finding global minima and calculating thermodynamic properties from basin-sampling. The GMIN homepage is located here.
Compiler compatibility
Tutorials
These tutorials are under development. Please correct any mistakes you find and, of course, feel free to add more!
- Compiling GMIN with CHARMM
- Adding a model to GMIN
- Global optimization of biomolecules using CHARMM
- Global optimization of biomolecules using AMBER9
- Global optimization of biomolecules using AMBER9 with Structural Restraints
- Optimization of ligand docking using AMBER9
- Restarting a GMIN run from a dump file
- Using the implicit membrane model IMM1
- Running a Go model with the AMHGMIN
Scripts
- makerestart: A bash script to automatically set up a GMIN restart run (for CHARMM users)